None
Note
This tutorial was generated from an IPython notebook that can be accessed from github.
Create 2D integer masks
In this tutorial we will show how to create 2D integer mask for arbitrary latitude and longitude grids.
Note
2D masks are good for plotting. However, to calculate weighted regional averages 3D boolean masks are more convenient. See the tutorial on 3D masks. See #226 how weighted regional averages can be calculated with 2D integer mask (this may also offer some speed gains).
Import regionmask and check the version:
import regionmask
regionmask.__version__
'0.12.1.post1.dev10+g9fd2ed6'
Load xarray and the tutorial data:
import numpy as np
import xarray as xr
xr.set_options(display_style="text", display_width=60)
<xarray.core.options.set_options at 0x7f13a9d2c170>
Creating a mask
Define a lon/ lat grid with a 1° grid spacing, where the points define the center of the grid.
lon = np.arange(-179.5, 180)
lat = np.arange(-89.5, 90)
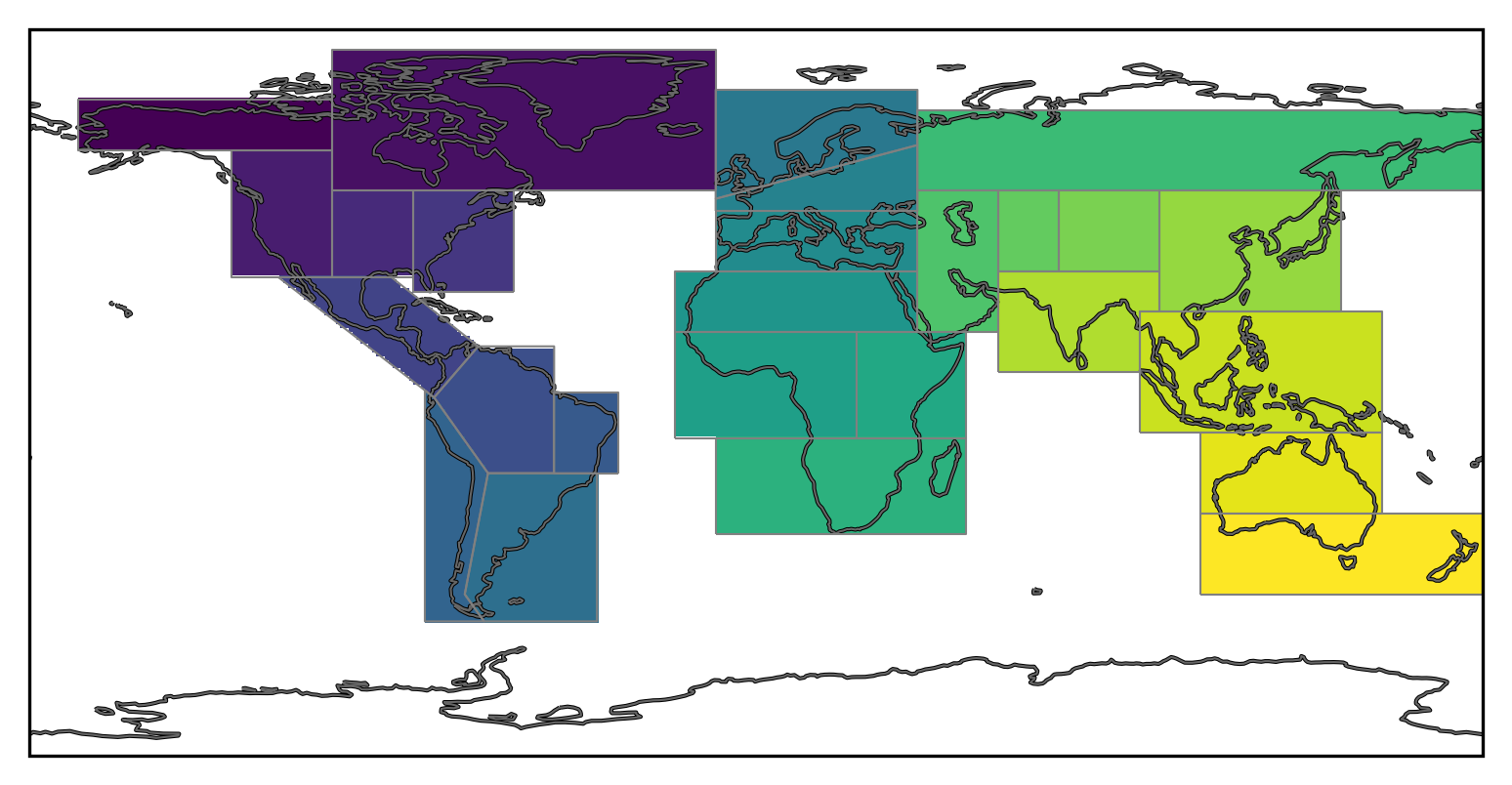
We will create a mask with the SREX regions (Seneviratne et al., 2012).
regionmask.defined_regions.srex
<regionmask.Regions 'SREX'>
Source: Seneviratne et al., 2012 (https://www.ipcc.ch/site/assets/uploads/2...
overlap: False
Regions:
1 ALA Alaska/N.W. Canada
2 CGI Canada/Greenl./Icel.
3 WNA W. North America
4 CNA C. North America
5 ENA E. North America
.. .. ...
22 EAS E. Asia
23 SAS S. Asia
24 SEA S.E. Asia
25 NAU N. Australia
26 SAU S. Australia/New Zealand
[26 regions]
The function mask determines which gridpoints lie within the polygon
making up the each region:
mask = regionmask.defined_regions.srex.mask(lon, lat)
mask
<xarray.DataArray 'mask' (lat: 180, lon: 360)> Size: 518kB
array([[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]])
Coordinates:
* lat (lat) float64 1kB -89.5 -88.5 ... 88.5 89.5
* lon (lon) float64 3kB -179.5 -178.5 ... 178.5 179.5
Attributes:
standard_name: region
flag_values: [ 1 2 3 4 5 6 7 8 9 10 11 12 ...
flag_meanings: ALA CGI WNA CNA ENA CAM AMZ NEB WSA S...mask is now a xarray.Dataset with shape lat x lon (if you
need a numpy array use mask.values). Gridpoints that do not fall in
a region are NaN, the gridpoints that fall in a region are encoded
with the number of the region (here 1 to 26).
We can now plot the mask:
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
f, ax = plt.subplots(subplot_kw=dict(projection=ccrs.PlateCarree()))
ax.coastlines()
regionmask.defined_regions.srex.plot(
ax=ax, add_label=False, line_kws=dict(lw=0.5, color="0.5")
)
mask.plot(ax=ax, transform=ccrs.PlateCarree(), add_colorbar=False)
<cartopy.mpl.geocollection.GeoQuadMesh at 0x7f13a18c4f20>
Working with a mask
masks can be used to select data in a certain region and to calculate regional averages - let’s illustrate this with a ‘real’ dataset:
airtemps = xr.tutorial.load_dataset("air_temperature")
The example data is a temperature field over North America. Let’s plot the first time step:
# choose a good projection for regional maps
proj = ccrs.LambertConformal(central_longitude=-100)
ax = plt.subplot(111, projection=proj)
airtemps.isel(time=1).air.plot.pcolormesh(ax=ax, transform=ccrs.PlateCarree())
ax.coastlines();

Conveniently we can directly pass an xarray object to the mask
function. It gets the longitude and latitude from the DataArray/
Dataset and creates the mask. Per default regionmask assumes the
longitude and latitude are called "lon" and "lat". If they have
another name, you can pass them individually,
e.g. region.mask(ds.longitude, ds.latitude).
mask = regionmask.defined_regions.srex.mask(airtemps)
Note
regionmask automatically detects whether the longitude needs to be wrapped around, i.e. if the regions extend from -180° E to 180° W, while the grid goes from 0° to 360° W as in our example:
lon = airtemps.lon.values
print(f"Grid extent: {lon.min():3.0f}°E to {lon.max():3.0f}°E")
bounds = regionmask.defined_regions.srex.bounds_global
print(f"Region extent: {bounds[0]:3.0f}°E to {bounds[2]:3.0f}°E")
Grid extent: 200°E to 330°E
Region extent: -168°E to 180°E
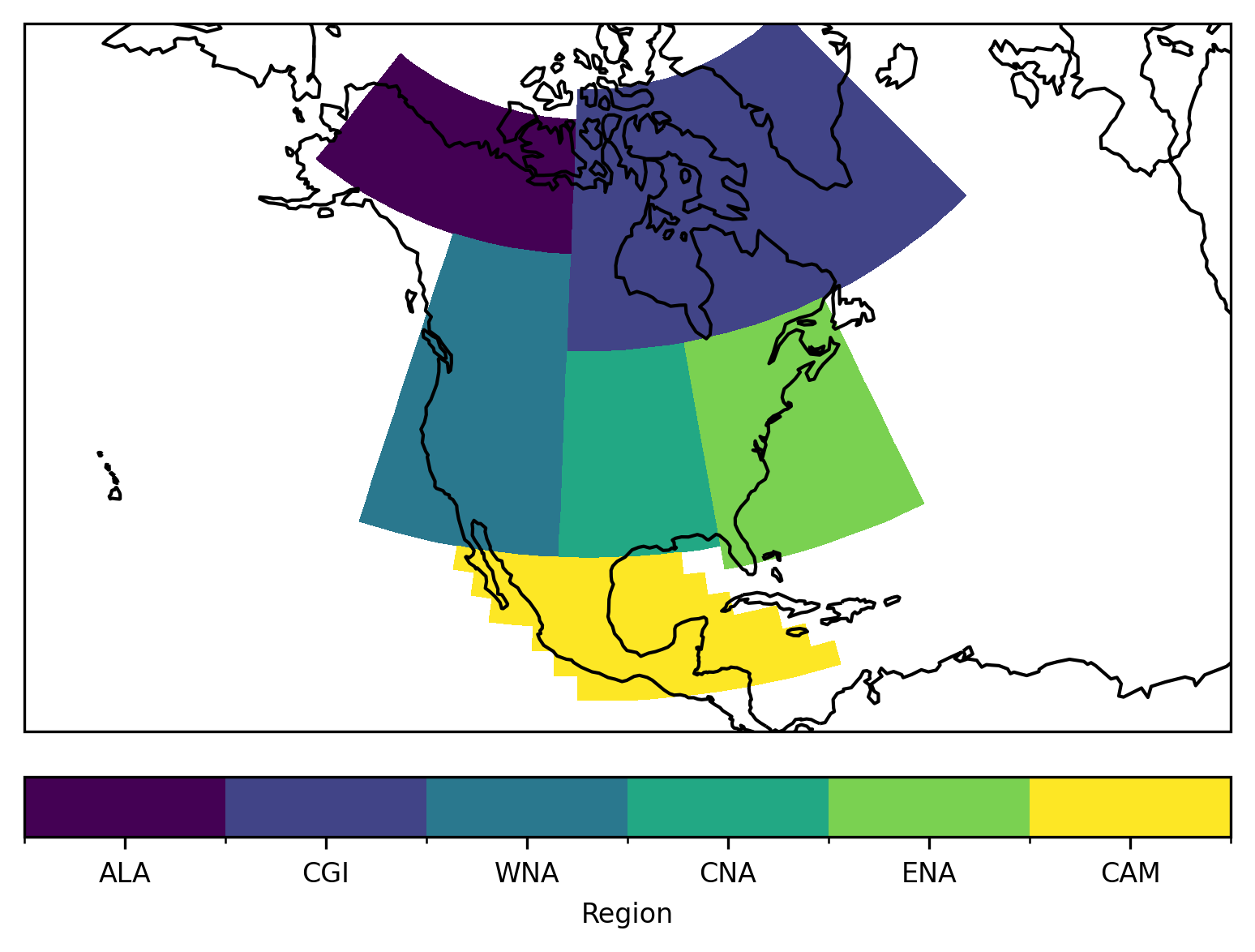
Let’s plot the mask of the regions:
proj = ccrs.LambertConformal(central_longitude=-100)
ax = plt.subplot(111, projection=proj)
low = mask.min()
high = mask.max()
levels = np.arange(low - 0.5, high + 1)
h = mask.plot.pcolormesh(
ax=ax, transform=ccrs.PlateCarree(), levels=levels, add_colorbar=False
)
# for colorbar: find abbreviations of all regions that were selected
reg = np.unique(mask.values)
reg = reg[~np.isnan(reg)]
abbrevs = regionmask.defined_regions.srex[reg].abbrevs
cbar = plt.colorbar(h, orientation="horizontal", fraction=0.075, pad=0.05)
cbar.set_ticks(reg)
cbar.set_ticklabels(abbrevs)
cbar.set_label("Region")
ax.coastlines()
# fine tune the extent
ax.set_extent([200, 330, 10, 75], crs=ccrs.PlateCarree())
Select a specific region - using cf_xarray
If cf_xarray
is installed we can select any individual region using the
flags saved
in the attributes (attrs) of the mask (new in regionmask 0.10.0):
mask.cf
Flag Variable:
Flag Meanings: ALA: 1
CGI: 2
WNA: 3
CNA: 4
ENA: 5
CAM: 6
Coordinates:
CF Axes: * X: ['lon']
* Y: ['lat']
Z, T: n/a
CF Coordinates: * longitude: ['lon']
* latitude: ['lat']
vertical, time: n/a
Cell Measures: area, volume: n/a
Standard Names: * latitude: ['lat']
* longitude: ['lon']
Bounds: n/a
Grid Mappings: n/a
Thus to select the region named “Central North America” (abbreviated “CNA”) we can do:
mask_CNA = mask.cf == "CNA"
# show a subset
mask_CNA.sel(lat=30, lon=slice(240, 260))
<xarray.DataArray 'mask' (lon: 9)> Size: 9B
array([False, False, False, False, False, False, False, True, True])
Coordinates:
lat float32 4B 30.0
* lon (lon) float32 36B 240.0 242.5 ... 257.5 260.0Warning
flag_meanings cannot contain spaces - they will be replaced by underscores:
mask_names = regionmask.defined_regions.srex.mask(airtemps, flag="names")
try:
mask_names.cf == "C. North America"
except ValueError:
print("error raised")
mask_names.cf == "C. North America".replace(" ", "_")
error raised
<xarray.DataArray 'mask' (lat: 25, lon: 53)> Size: 1kB
array([[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]])
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 ... 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 ... 327.5 330.0Select a specific region - without cf_xarray
We first need to find out which number the region ‘C. North America’ corresponds to:
CNA_index = regionmask.defined_regions.srex.map_keys("C. North America")
CNA_index
4
mask_CNA = mask == CNA_index
Mask out a region
To replace all grid points outside of CNA we use the where method of
xr.Dataset
(documentation),
which filters elements from this object according to a condition:
airtemps_CNA = airtemps.where(mask_CNA)
Check everything went well by repeating the first plot with the selected region:
# choose a good projection for regional maps
proj = ccrs.LambertConformal(central_longitude=-100)
ax = plt.subplot(111, projection=proj)
regionmask.defined_regions.srex[["CNA"]].plot(ax=ax, add_label=False)
airtemps_CNA.isel(time=1).air.plot.pcolormesh(ax=ax, transform=ccrs.PlateCarree())
ax.coastlines();
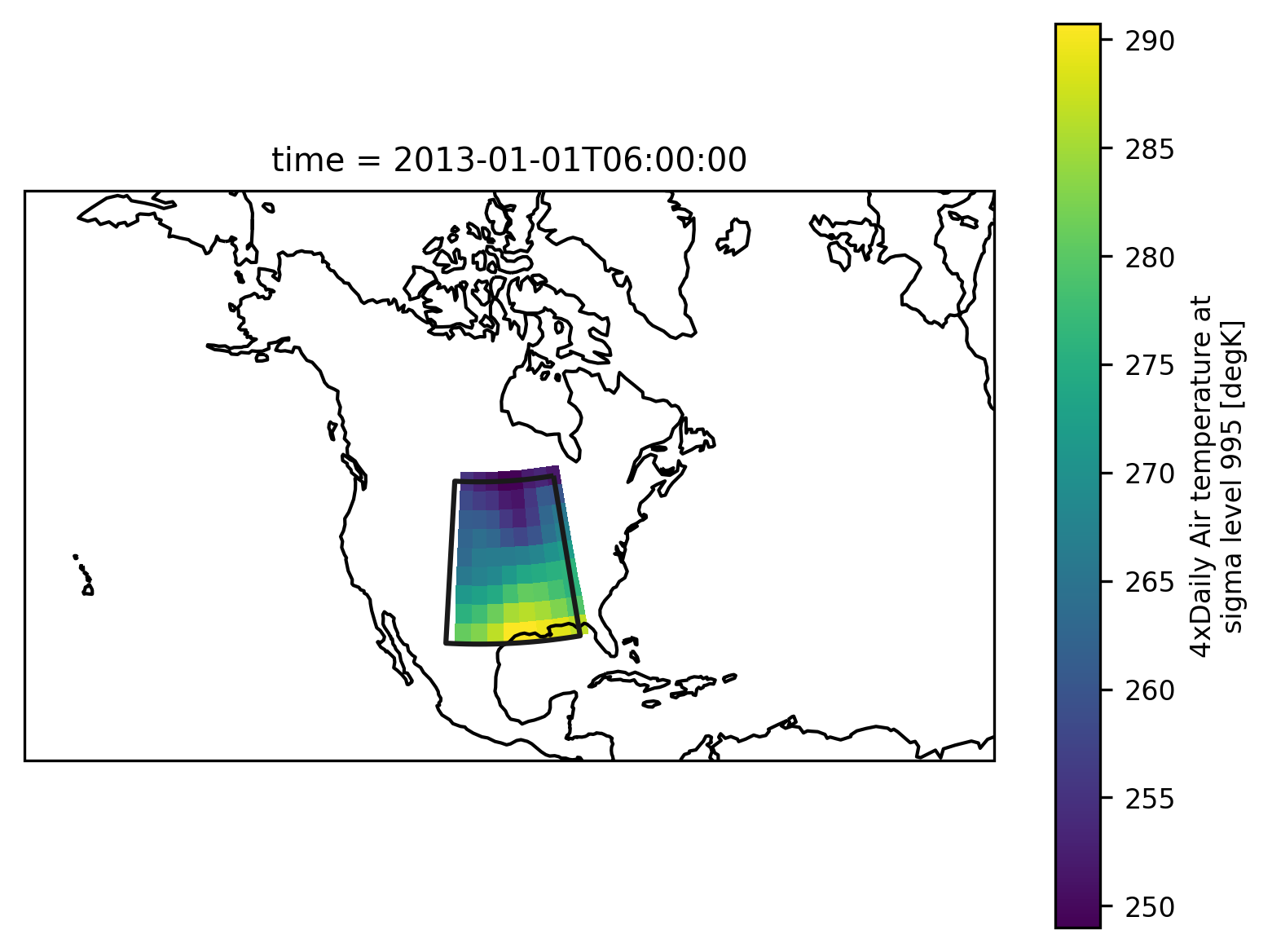
Looks good - with this we can calculate the region average.
Calculate weighted regional average
From version 0.15.1 xarray includes a function to calculate the weighted
mean - we use cos(lat) as proxy of the grid cell area
Note
It is better to use a model’s original grid cell area (e.g. areacella). cos(lat) works reasonably well for regular lat/ lon grids. For irregular grids (regional models, ocean models, …) it is not appropriate.
weights = np.cos(np.deg2rad(airtemps.lat))
ts_airtemps_CNA = airtemps_CNA.weighted(weights).mean(dim=("lat", "lon")) - 273.15
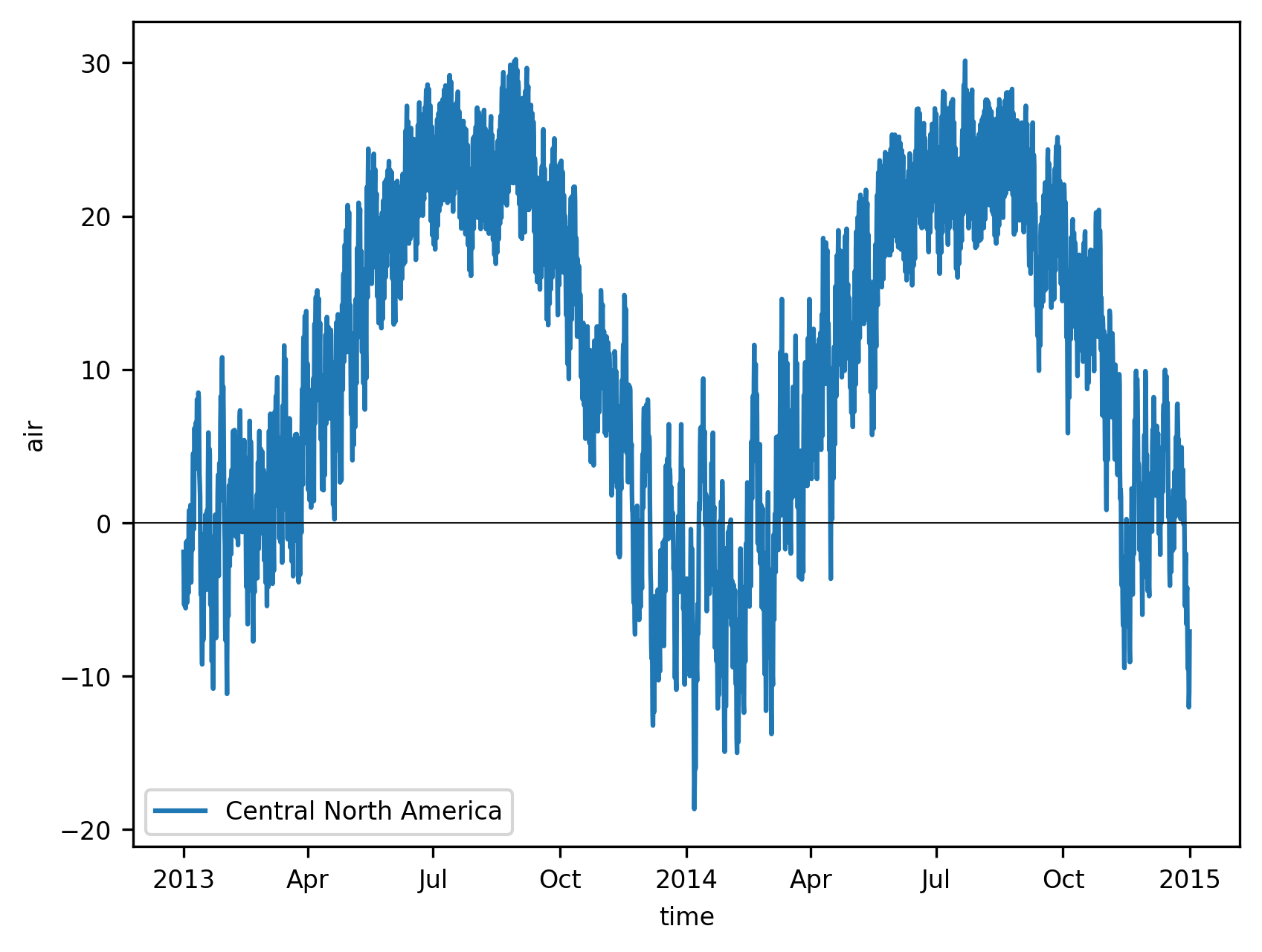
We plot the resulting time series:
f, ax = plt.subplots()
ts_airtemps_CNA.air.plot.line(ax=ax, label="Central North America")
ax.axhline(0, color="0.1", lw=0.5)
plt.legend();
To get the regional average for each region you would need to loop over them. However, it’s easier to use a 3D mask.
Calculate regional statistics using groupby
Warning
Using groupby offers some convenience and is faster than using where and a loop. However, xarray does currently not natively support to combine groupby with weighted (pydata/xarray#3937), see #226 for a workaround.
Overall, I recommend working with a 3D mask.
# you can group over all integer values of the mask
airtemps_all = airtemps.groupby(mask).mean()
airtemps_all
<xarray.Dataset> Size: 164kB
Dimensions: (mask: 6, time: 2920)
Coordinates:
* time (time) datetime64[ns] 23kB 2013-01-01 ... 20...
* mask (mask) float64 48B 1.0 2.0 3.0 4.0 5.0 6.0
Data variables:
air (mask, time) float64 140kB 254.0 ... 294.9
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridd...However, groupby is the way to go when calculating a (unweighted)
regional median:
# you can group over all integer values of the mask
airtemps_reg_median = airtemps.groupby(mask).median()
airtemps_reg_median.isel(time=0)
<xarray.Dataset> Size: 104B
Dimensions: (mask: 6)
Coordinates:
time datetime64[ns] 8B 2013-01-01
* mask (mask) float64 48B 1.0 2.0 3.0 4.0 5.0 6.0
Data variables:
air (mask) float64 48B 249.8 250.6 ... 280.4 296.0
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridd...Multidimensional coordinates
Regionmask can also handle mutltidimensional longitude/ latitude grids (e.g. from a regional climate model). As xarray provides such an example dataset, we will use it to illustrate it. See also in the xarray documentation.
Load the tutorial data:
rasm = xr.tutorial.load_dataset("rasm")
The example data is a temperature field over the Northern Hemisphere. Let’s plot the first time step:
# choose a projection
proj = ccrs.NorthPolarStereo()
ax = plt.subplot(111, projection=proj)
ax.set_global()
rasm.isel(time=1).Tair.plot.pcolormesh(
ax=ax, x="xc", y="yc", transform=ccrs.PlateCarree()
)
# add the abbreviation of the regions
regionmask.defined_regions.srex[[1, 2, 11, 12, 18]].plot(
ax=ax, add_coastlines=False, label="abbrev"
)
ax.set_extent([-180, 180, 43, 90], ccrs.PlateCarree())
ax.coastlines();
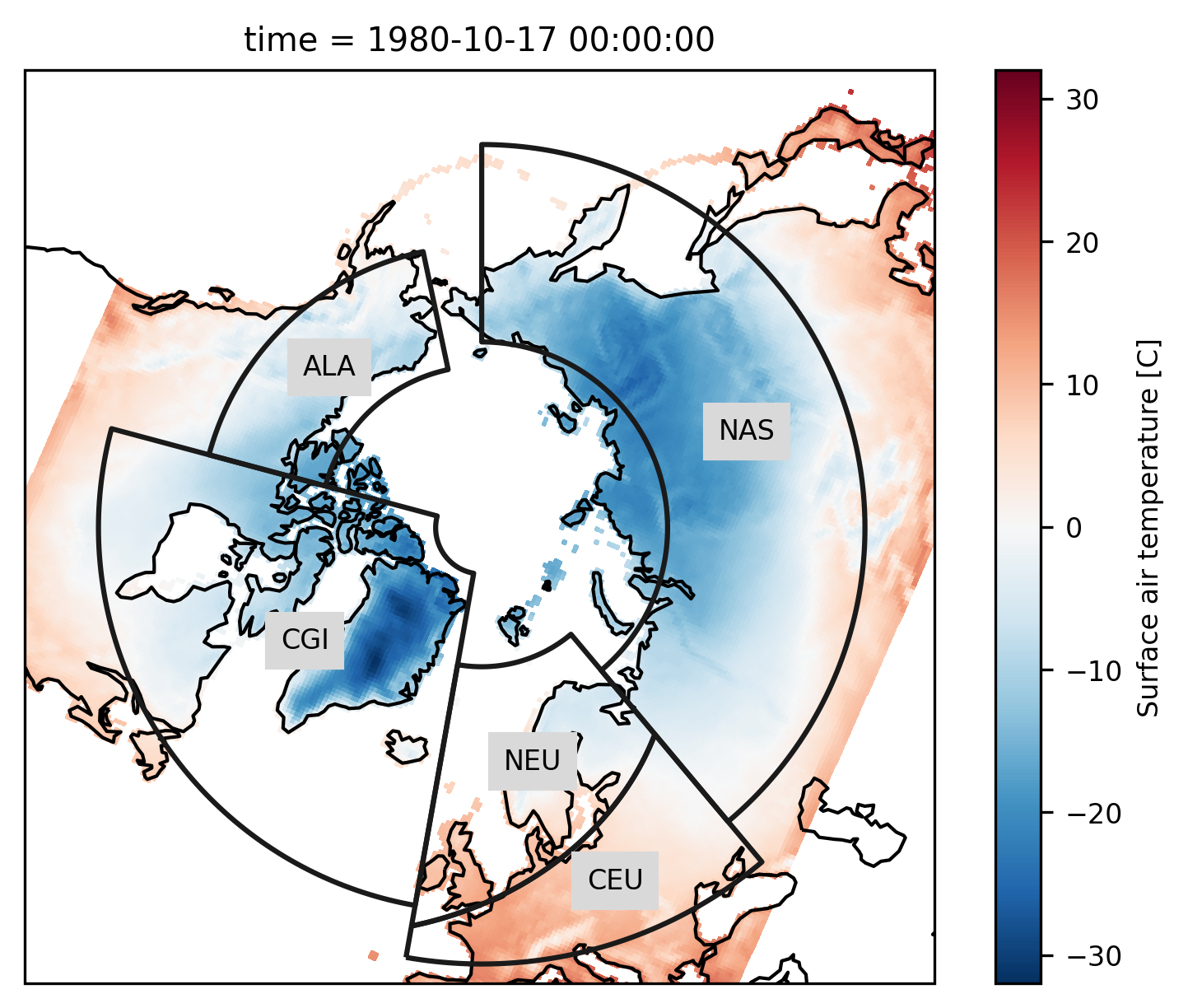
Again we pass the xarray object to regionmask. Here, cf_xarray is used
to detect rasm.xc and rasm.yc as longitude and latitude
coordinates (new in regionmask 0.10.0). Without cf_xarray we would need
to pass the coordinates explicitly
(i.e. srex.mask(rasm.xc, rasm.yc):
mask = regionmask.defined_regions.srex.mask(rasm)
mask
<xarray.DataArray 'mask' (y: 205, x: 275)> Size: 451kB
array([[nan, nan, nan, ..., 5., 5., 5.],
[nan, nan, nan, ..., 5., 5., 5.],
[nan, nan, nan, ..., 5., 5., 5.],
...,
[24., 24., 24., ..., 14., 14., 14.],
[24., 24., 24., ..., 14., 14., 14.],
[24., 24., 24., ..., 14., 14., 14.]])
Coordinates:
xc (y, x) float64 451kB 189.2 189.4 ... 16.91
yc (y, x) float64 451kB 16.53 16.78 ... 27.51
Dimensions without coordinates: y, x
Attributes:
standard_name: region
flag_values: [ 1 2 3 4 5 11 12 13 14 18 19 20 ...
flag_meanings: ALA CGI WNA CNA ENA NEU CEU MED SAH N...We want to select the region ‘NAS’ (Northern Asia).
Select using where
Using the cf_xarray accessor (from regionmask 0.10.0) we can directly use the name of the region (otherwise we need to select by the the number of the region):
rasm_NAS = rasm.where(mask.cf == "NAS")
Check everything went well by repeating the first plot with the selected region:
# choose a projection
proj = ccrs.NorthPolarStereo()
ax = plt.subplot(111, projection=proj)
ax.set_global()
rasm_NAS.isel(time=1).Tair.plot.pcolormesh(
ax=ax, x="xc", y="yc", transform=ccrs.PlateCarree()
)
# add the abbreviation of the regions
regionmask.defined_regions.srex[["NAS"]].plot(
ax=ax, add_coastlines=False, label="abbrev"
)
ax.set_extent([-180, 180, 45, 90], ccrs.PlateCarree())
ax.coastlines();
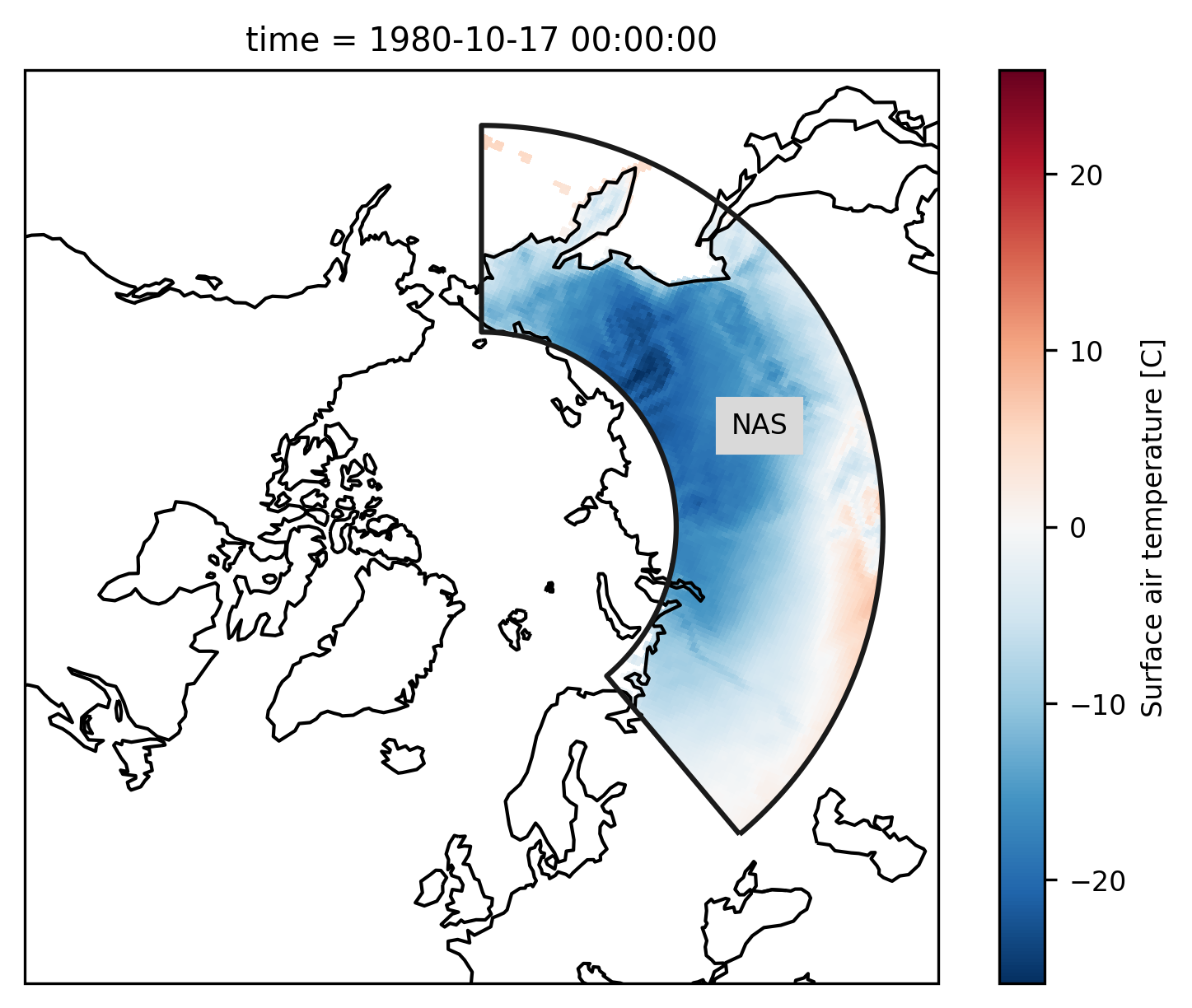
References
Special Report on Managing the Risks of Extreme Events and Disasters to Advance Climate Change Adaptation (SREX, Seneviratne et al., 2012)