None
Note
This tutorial was generated from an IPython notebook that can be accessed from github.
Working with geopandas (shapefiles)
regionmask includes support for regions defined as geopandas
GeoDataFrame. These are often shapefiles, which can be opened in the
formats .zip, .shp, .geojson etc. with
geopandas.read_file(url_or_path).
There are two possibilities:
Directly create a mask from a geopandas GeoDataFrame or GeoSeries using
mask_geopandasormask_3D_geopandas.Convert a GeoDataFrame to a
Regionsobject (regionmask’s internal data container) usingfrom_geopandas.
As always, start with the imports:
import cartopy.crs as ccrs
import geopandas as gp
import matplotlib.patheffects as pe
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pooch
import regionmask
regionmask.__version__
'0.12.1.post1.dev10+g9fd2ed6'
Opening an example shapefile
The U.S. Geological Survey (USGS) offers a shapefile containing the outlines of continens [1]. We use the library pooch to locally cache the file:
file = pooch.retrieve(
"https://pubs.usgs.gov/of/2006/1187/basemaps/continents/continents.zip", None
)
continents = gp.read_file("zip://" + file)
display(continents)
Downloading data from 'https://pubs.usgs.gov/of/2006/1187/basemaps/continents/continents.zip' to file '/home/docs/.cache/pooch/7dd514faeaa71efe73294dece9245e99-continents.zip'.
SHA256 hash of downloaded file: af0ba524a62ad31deee92a9700fc572088c2b93a39ba66f320677dd8dacaaaaf
Use this value as the 'known_hash' argument of 'pooch.retrieve' to ensure that the file hasn't changed if it is downloaded again in the future.
CONTINENT geometry
0 Asia MULTIPOLYGON (((93.27554 80.26361, 93.31304 80...
1 North America MULTIPOLYGON (((-25.28167 71.39166, -25.32889 ...
2 Europe MULTIPOLYGON (((58.06138 81.68776, 57.98055 81...
3 Africa MULTIPOLYGON (((0.69465 5.77337, 0.66667 5.803...
4 South America MULTIPOLYGON (((-81.71306 12.49028, -81.72014 ...
5 Oceania MULTIPOLYGON (((-177.39334 28.18416, -177.3958...
6 Australia MULTIPOLYGON (((142.27997 -10.26556, 142.21053...
7 Antarctica MULTIPOLYGON (((51.80305 -46.45667, 51.72139 -...
Create a mask from a GeoDataFrame
mask_geopandas and mask_3D_geopandas allow to directly create a
mask from a GeoDataFrame or GeoSeries:
lon = np.arange(-180, 180)
lat = np.arange(-90, 90)
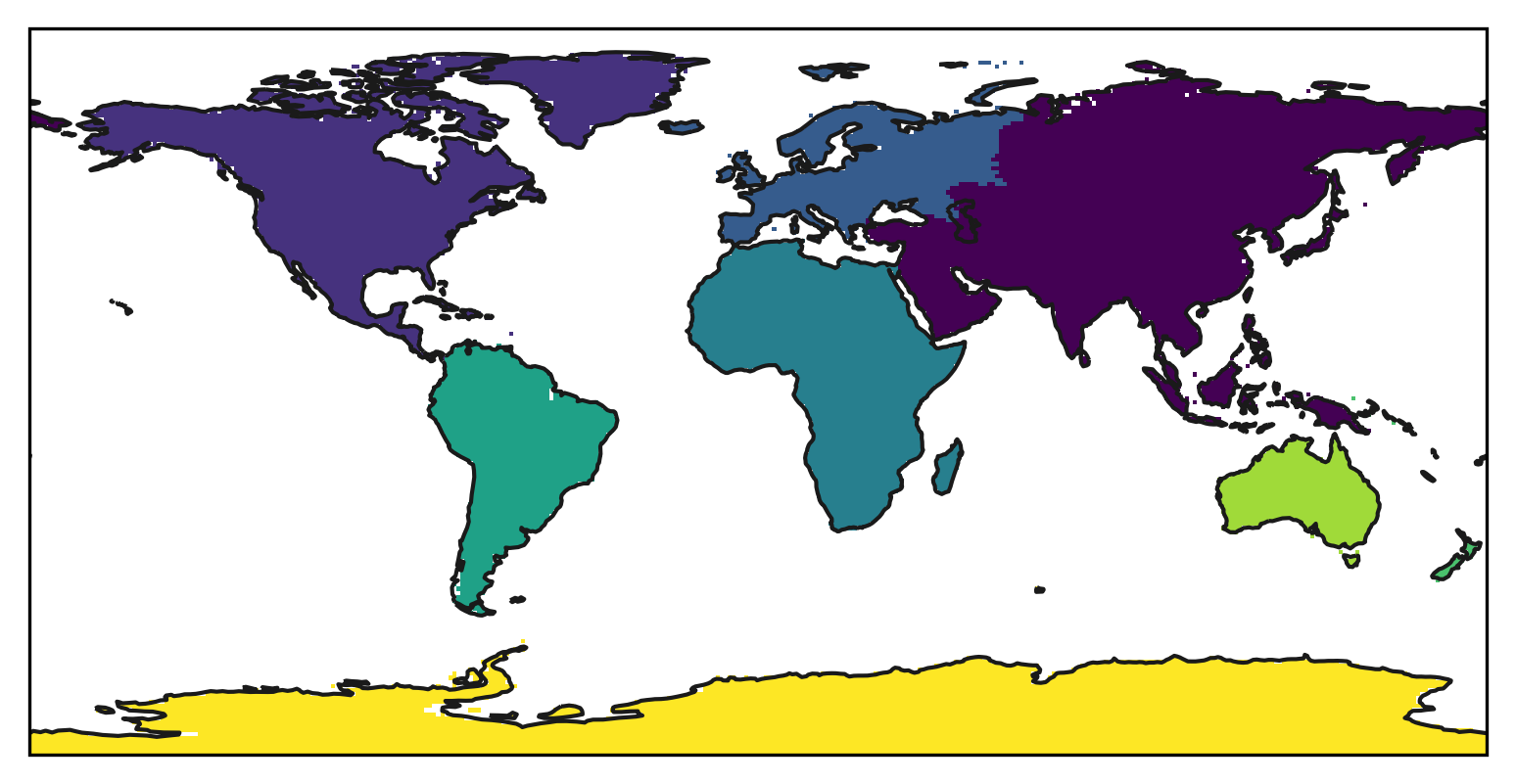
mask = regionmask.mask_geopandas(continents, lon, lat)
Let’s plot the new mask:
f, ax = plt.subplots(subplot_kw=dict(projection=ccrs.PlateCarree()))
mask.plot(
ax=ax,
transform=ccrs.PlateCarree(),
add_colorbar=False,
)
ax.coastlines(color="0.1");
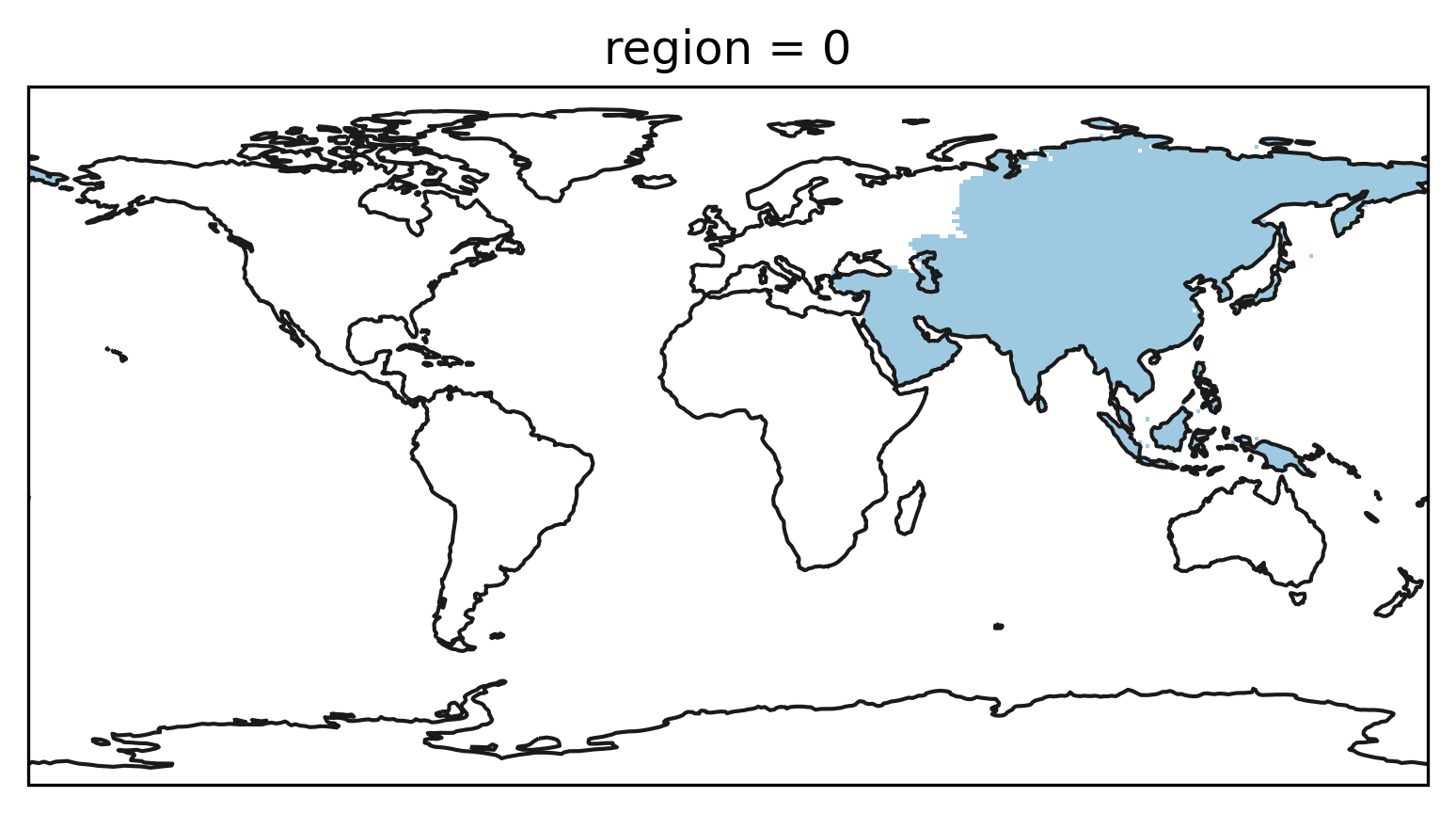
Similarly a 3D boolean mask can be created from a GeoDataFrame:
mask_3D = regionmask.mask_3D_geopandas(continents, lon, lat)
and plotted:
from matplotlib import colors as mplc
cmap1 = mplc.ListedColormap(["none", "#9ecae1"])
f, ax = plt.subplots(subplot_kw=dict(projection=ccrs.PlateCarree()))
mask_3D.sel(region=0).plot(
ax=ax,
transform=ccrs.PlateCarree(),
add_colorbar=False,
cmap=cmap1,
)
ax.coastlines(color="0.1");
Note
Set regionmask.mask_3D_geopandas(..., overlap=True) if some of the regions overlap. See the tutorial on overlapping regions for details.
2. Convert GeoDataFrame to a Regions object
Creating a Regions object with regionmask.from_geopandas
requires a GeoDataFrame:
continents_regions = regionmask.from_geopandas(continents)
continents_regions
<regionmask.Regions 'unnamed'>
overlap: None
Regions:
0 r0 Region0
1 r1 Region1
2 r2 Region2
3 r3 Region3
4 r4 Region4
5 r5 Region5
6 r6 Region6
7 r7 Region7
[8 regions]
This creates default names ("Region0", …, "RegionN") and
abbreviations ("r0", …, "rN").
However, it is often advantageous to use columns of the GeoDataFrame as
names and abbrevs. If no column with abbreviations is available, you can
use abbrevs='_from_name', which creates unique abbreviations using
the names column.
continents_regions = regionmask.from_geopandas(
continents, names="CONTINENT", abbrevs="_from_name", name="continent"
)
continents_regions
<regionmask.Regions 'continent'>
overlap: None
Regions:
0 Asi Asia
1 NorAme North America
2 Eur Europe
3 Afr Africa
4 SouAme South America
5 Oce Oceania
6 Aus Australia
7 Ant Antarctica
[8 regions]
Note
Set overlap=True if some of the regions overlap. See the tutorial on overlapping regions for details.
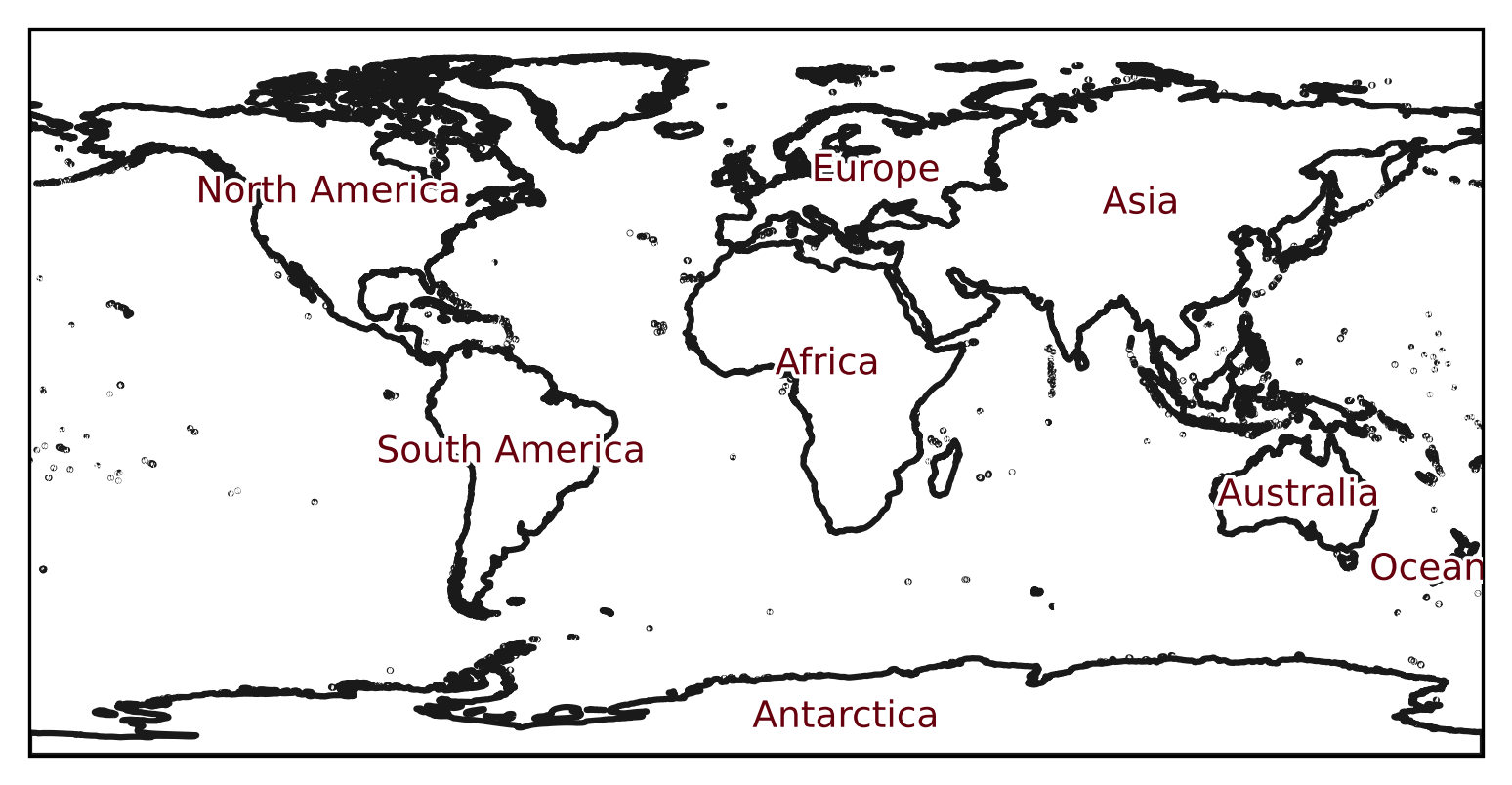
As usual the newly created Regions object can be plotted on a world
map:
text_kws = dict(
bbox=dict(color="none"),
path_effects=[pe.withStroke(linewidth=2, foreground="w")],
color="#67000d",
fontsize=9,
)
continents_regions.plot(label="name", add_coastlines=False, text_kws=text_kws);
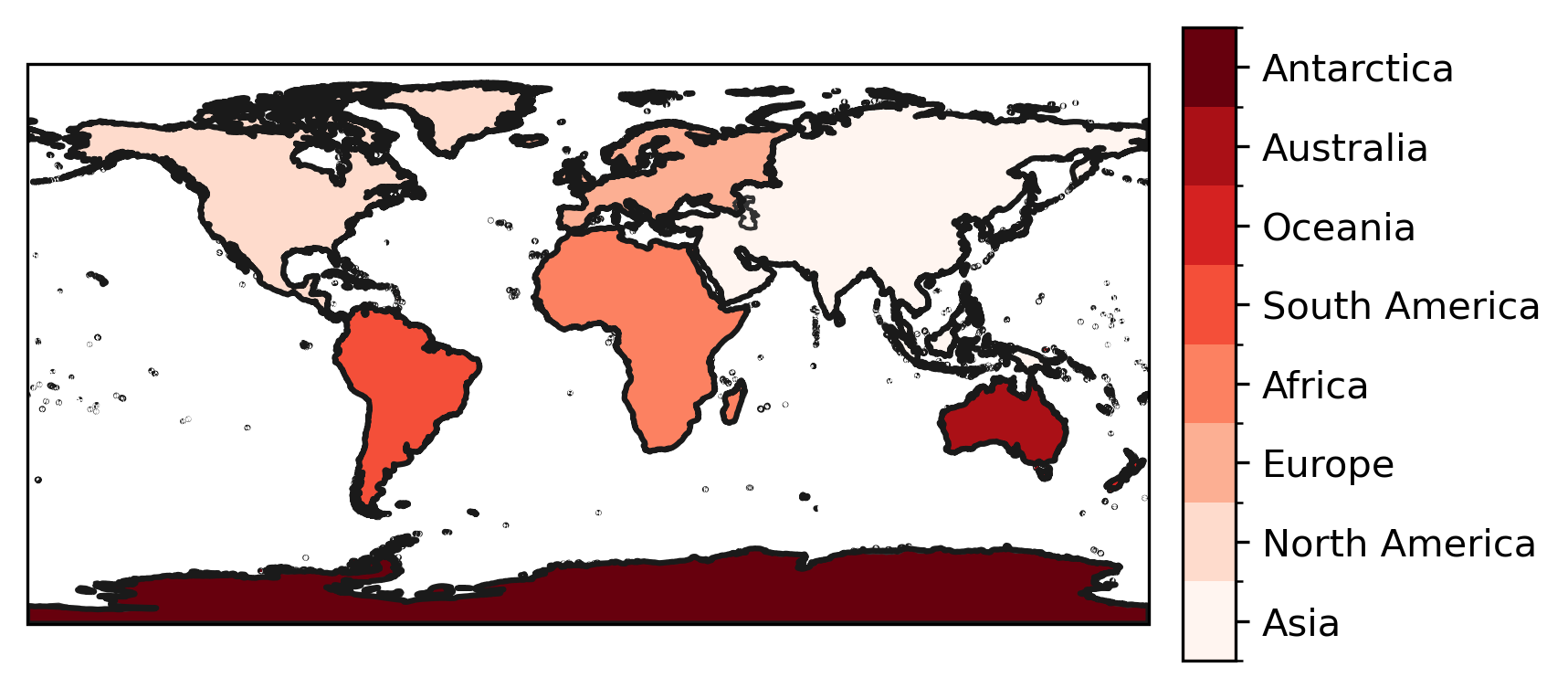
And to create mask a mask for arbitrary latitude/ longitude grids:
lon = np.arange(0, 360)
lat = np.arange(-90, 90)
mask = continents_regions.mask(lon, lat)
which can then be plotted
f, ax = plt.subplots(subplot_kw=dict(projection=ccrs.PlateCarree()))
h = mask.plot(
ax=ax,
transform=ccrs.PlateCarree(),
cmap="Reds",
add_colorbar=False,
levels=np.arange(-0.5, 8),
)
cbar = plt.colorbar(h, shrink=0.625, pad=0.025, aspect=12)
cbar.set_ticks(np.arange(8))
cbar.set_ticklabels(continents_regions.names)
ax.coastlines(color="0.2")
continents_regions.plot_regions(add_label=False);
References
[1] Environmental Systems Research , Inc. (ESRI), 20020401, World Continents: ESRI Data & Maps 2002, Environmental Systems Research Institute, Inc. (ESRI), Redlands, California, USA.